We propose a method to apply transfer learning to scRNA-Seq data that enables us to transfer knowledge from a relatively well-annotated and large source dataset to a smaller unannotated target dataset. A graphical representation of the method can be found in Fig. 1B. The method is based on a transfer learning step, that modifies the target dataset to incorporate knowledge gained from the well-annotated source dataset. The newly constructed target dataset can then be analyzed with a clustering algorithm to obtain an improved clustering compared to applying that same method to the target without any transfer learning procedure or a simple concatenation of source and target.
The following sections describe the method in more detail and specify the experimental setup of performance assessments on generated synthetic data, controlled real data and a real-world application of two independent datasets.
Transfer learning for scRNA-seq clustering
There exists a well-known source dataset Xsrc with scRNA-Seq data from nsrc cells and g genes for which we have in-depth knowledge about the clustering structure (i.e. ground truth labels ({y}^{src}in {Re }^{{n}_{src}})) and a target dataset Xtrg of ntrg cells and g genes which we want to enhance given the information in Xsrc and ysrc before clustering into k groups of cells.
The basic underlying idea of the proposed method is to factorize the source dataset into a gene independent part (of size k × nsrc) and a data size independent part (of size g × k) and to use the latter – which is often called a dictionary since it does not depend on the number of cells nsrc and can thus be used to translate between datasets – to modify the target dataset accordingly.
More specifically, the novel approach, based on non-negative matrix factorization (NMF), can be derived in the following steps:
- 1.
We use NMF73,74 of our source data ({X}_{src}in {Re }^{gtimes {n}_{src}}) to learn a dictionary ({H}_{src}in {Re }^{gtimes k}) and a data matrix ({W}_{src}in {Re }^{ktimes {n}_{src}}) while regularizing the denseness of the results with an elastic net75:
$$begin{array}{rcl}{H}_{src},{W}_{src} & = & argmi{n}_{W,H}(frac{1}{2}{||{X}_{src}-HW||}_{Fro}^{2}+alpha lambda ({||vec(H)||}_{1}+{||vec(W)||}_{1})\ & & +,frac{alpha }{2}(1-lambda )({||H||}_{Fro}^{2}+{||W||}_{Fro}^{2}))end{array}$$
Here, λ is the elastic net mixing parameter controlling the combination of L1 and L2 regularization and α is the corresponding penalty multiplier.
As an initial starting point ({W}_{src}^{ast }) for Wsrc we provide a one-hot-encoding of the given cluster labels ysrc, where a non-zero entry in the j-th row of column i in ({W}_{src}^{ast }) indicates that cell i is a member of cluster j.
- 2.
Given the learned dictionary ({H}_{src}in {Re }^{gtimes k}) from step (1) and assuming the genes in source and target data correspond, we now transfer knowledge from the source to the target dataset through the dictionary by learning a target data matrix ({W}_{trg}in {Re }^{ktimes {n}_{trg}}):
$${W}_{trg}=argmi{n}_{W}(frac{1}{2}{||{X}_{trg}-{H}_{src}W||}_{Fro}^{2})$$
- 3.
To enable domain adaptation for different levels of cell type overlap between the two datasets we now construct a new target dataset ({X}_{trg}^{new}) based on a convex combination of a reconstructed target dataset ({H}_{src}{W}_{trg}^{text{‘}}) and its original version Xtrg:
$${X}_{trg}^{new}=theta {H}_{src}{W}_{trg}^{text{‘}}+(1-theta ){X}_{trg},{rm{with}},0le theta le 1$$
θ is a mixture parameter indicating how strongly knowledge from the source dataset should be transferred into the newly constructed target dataset. High values of θ indicate a strong influence of the source dataset on the modified dataset and low values cause the new dataset to be more similar to its original version.
The target clustering matrix ({W}_{trg}^{text{‘}}in {{0,1}}^{ktimes {n}_{trg}}) is a simplified version of Wtrg with ones at the positions of all column-wise maxima and zeros elsewhere, i.e.
$${({W}_{trg}^{text{‘}})}_{li}={1}_{[(argma{x}_{lin {1,ldots ,k}}{({W}_{trg})}_{li})=l]}forall i,l$$
Using ({W}_{trg}^{text{‘}}) instead of Wtrg represents reducing the information in this matrix to potential cluster memberships of the target cells which is appropriate considering the task at hand. To this end, a number of different approaches were implemented (e.g. leaving Wtrg as it is or optimizing it in an additional training step), but it was found that taking the simplified version as described above performed best and most consistently for all scenarios under investigation.
- 4.
The newly derived dataset ({X}_{trg}^{new}) can be used as input for a clustering method. We are using single-cell consensus clustering (SC3)37 as an exemplary clustering method that is commonly used to solve scRNA-Seq clustering problems. See Supplementary Section 1.1. for a detailed description of SC3.
Please note that the proposed method does not inherently depend on the number of samples in each dataset and can technically (even though not studied in this work) be used to transfer knowledge from datasets of any size, not just from a source that is larger than the target.
Selecting θ and other free parameters
The mixture parameter θ dictates how much the newly constructed target dataset should be changed by the information in the source dataset. θ is automatically chosen via an unsupervised assessment of the clustering quality through Kernel Target Alignment (KTA) scores76 which measure the similarity of kernels. The whole transfer learning and clustering procedure (steps 1–4) is applied with a number of values for θ within a pre-specified range and the KTA score between the linear kernel of the mixed dataset ({X}_{trg}^{new})over the cells and the linear kernel of the cell type labels predicted by subsequent SC3 clustering is calculated. The parameter value yielding the optimal KTA score is chosen for the final result and can give an indication on the transferability between source and target data. An investigation of the mixture parameter of the transfer learning approach and its automatic selection process based on KTA scores is given in Supplementary Section 2.4. where the correlation of the unsupervised KTA scores and their supervised counterpart, the Adjusted Rand Indices (ARIs) are examined. Other free parameters, e.g. the elastic net parameters λ and α, were chosen based on results from the simulated data. See Supplementary Sections 2.2.,3.2. and 4.2. for details.
Pre-processing
Three steps for pre-processing scRNA-Seq data were applied:
-
Cell filter: Remove all cells containing fewer than xgenes genes with expression > xexpression.
-
Gene filter: Remove ubiquitous genes that are expressed in almost all cells (i.e. with expression > xexpression in at least xcells% of cells) and rare genes that are not expressed in almost all cells (i.e. with expression < xexpression in at least xcells% of cells).
-
Log-transformation: Log-transform the expression matrix after adding a pseudo-count of 1.
All free pre-processing parameters should be selected by the user based on an inspection of the data, i.e. expression histograms of both source and target dataset. The specific parameter values chosen for the datasets in this work can be found in Supplementary Sections 2.1.,3.1. and 4.1. The corresponding expression histograms can be seen in Supplementary Figs. S5–S7. Pre-processing was performed once for all datasets (source and target separately) before the different clustering methods (i.e. transfer learning or baseline methods) were performed.
NMF clustering in the absence of source labels
If no reliable cluster labels are available for the source dataset ({X}_{src}in {Re }^{gtimes {n}_{src}}), one can choose to generate those labels via NMF clustering73,74 and proceed as if they were the real labels ysrc. This basically consists of learning a dictionary ({H}_{src}in {Re }^{gtimes k}) and a data matrix ({W}_{src}in {Re }^{ktimes {n}_{src}}) as described above in step 1 and selecting the cluster memberships based on the column wise maxima of Wsrc, i.e. ({y}_{i}^{src}=argma{x}_{lin {1,ldots ,k}}{({W}_{src})}_{li}).
Instead of learning the source labels through NMF clustering one could also avoid providing an initial starting point ({W}_{src}^{ast }) for Wsrc when learning the dictionary Hsrc and the data matrix Wsrc.
Baseline methods and performance metrics
For assessing the quality of our unsupervised domain adaptation solution, we are interested in investigating the change of clustering accuracy of the target dataset. As baseline methods we implement the original SC3 clustering method on the target dataset alone (TargetCluster) and on the concatenated dataset of source and target (ConcatenateCluster). For a detailed description and a visualization of the baseline methods please see Supplementary Section 1.2 and Supplementary Fig. S2.
As a supervised performance metric we used the Adjusted Rand Index (ARI)77 comparing the transfer learning results (TransferCluster) and the baseline labels with the known clustering labels (known perfectly in the case of the simulated data and retrieved from the original publication in the case of the real data). ARI scores are computed only on the target data, even in the case of ConcatenateCluster, where labels are computed for both source and target cells.
Simulation of source and target single-cell datasets
To test the applicability of our method we first use it on simulated count level scRNA-Seq data from a defined hierarchical set of clusters that represent the different cell types present in a tissue or biofluid.
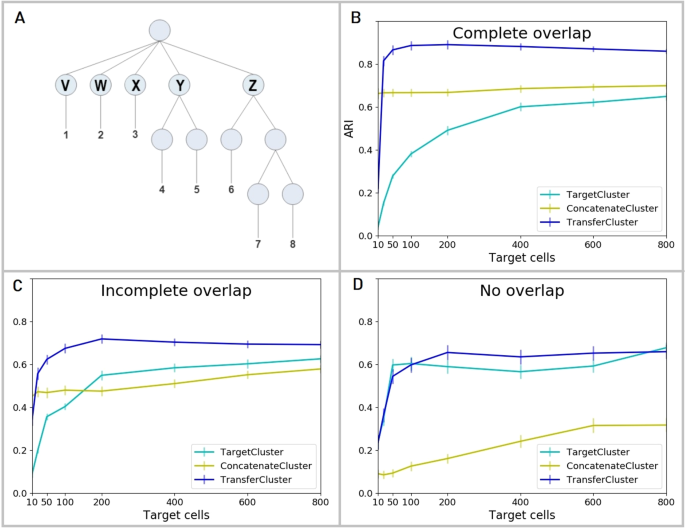
Figure 2A shows a graphical representation of the hierarchical clustering structure used to generate the simulated data. Each generated dataset consisted of eight clusters of cells (1–8) deriving from five top level clusters (V – Z) that share a common background distribution of gene expression levels and some proportion of genes differentially expressed between them.
scRNA-Seq simulation data description and results. (A) Count level single-cell RNA-Seq data is simulated according to a pre-defined hierarchical clustering structure with eight cell clusters (1-8) that are derived from five top level clusters (V – Z). Generated datasets are individually split up by randomly assigning the top node clusters V – Z to source or target. Three different settings are considered: 1. both source and target data contain cells from all top node clusters V – Z (Complete overlap), 2. three randomly selected top node clusters V – Z are chosen as common to both source and target, the other two are assigned to either one of source and target (Incomplete overlap) or 3. cells from two of the top node clusters form the target dataset and cells from the other three form the source (No overlap). (B) Clustering performances of the baseline methods, TargetCluster (clustering on the target dataset alone) and ConcatenateCluster (concatenating and clustering source and target data simultaneously), and the transfer learning approach (TransferCluster) when the clustering structures of source and target data are identical (Complete overlap). (C) Clustering performances of the baseline methods, TargetCluster and ConcatenateCluster and the transfer learning approach (TransferCluster) for an incomplete overlap between the cell clusters in source and target data (Incomplete overlap). (D) Clustering performances of the baseline methods, TargetCluster and ConcatenateCluster and the transfer learning approach (TransferCluster) for a setting with two exclusive target and three exclusive source top nodes and no cell types that appear in both sets (No overlap). Please note, that due to the sampling procedures described above, the number of top level nodes in the target datasets decreases from 5 in (B) to 4 in (C) and 2 in (D) and hence the performance of TargetCluster improves from (B–D). 95% confidence intervals are shown.
An outline of the data generation procedure is given here; the full code is provided in https://github.com/nicococo/scRNA/blob/master/scRNA/simulation.py. First we generate the number of cells in each sub-cluster using a Dirichlet distribution with a concentration parameter of 10. Then we define a common background distribution of gene expression levels sampled from a gamma distribution with shape 2 and rate 0.1. For each cluster in turn we randomly select 10–40 % of genes to be differentially expressed relative to the background. The difference in expression, expressed as a log2 fold change, for each such gene is sampled from a normal distribution with mean 1 and standard deviation 0.5. For clusters that are not themselves top level clusters (clusters 4–8) this process continues recursively with further expression differences generated for each sub-cluster using the parent cluster as the new background until the final clusters are reached. Finally, we generate count level data by applying a small amount of random normally distributed noise to the expression levels of each cell and then sampling the per gene counts from a negative binomial distribution with dispersion 0.1. The resulting datasets contain cells with a median count of 215 500 reads per cell. Please see Supplementary Section 2.3 and Supplementary Fig. S3 for details.
Once count level data is generated for the entire dataset we split it into target and source datasets with different sets of cells according to the cluster structure and the relationship between the target and source. Here, we consider three such relationships that reflect three possible experimental scenarios:
-
Cells in the target and source are randomly sampled from the same underlying tissue or biofluid and hence contain cells from all top node clusters V–Z.
-
Certain clusters are specified to be only present in the source and some to be only present in the target, the remaining clusters are present in both target and source. Three randomly selected top node clusters V – Z are chosen as common to both source and target, the other two are assigned to either one of source and target.
-
The cells in the target and source are drawn from completely non-overlapping clusters. In this scenario transfer learning is not expected to be successful. Cells from two of the top node clusters form the target dataset and cells from the other three form the source.
The genes measured in source and target are always the same and the top nodes are randomly assigned to either source or target for each repetition of the data generation process. 100 sets of simulated data were generated for each of the three settings simulating the expression levels of 10 000 genes in 1800 cells. 1000 cells were assigned to the source dataset and the set of 800 target cells was downsampled (i.e. 10, 50, 100, 200, 400, 600, 800 target cells) to investigate the performance of the transfer learning approach and its corresponding baseline methods when applied to datasets of varying sizes.
Subsampling source and target data from a single dataset
Following the analysis of simulated data, we subsequently examined a real scRNA-Seq dataset. By subsampling both source and target dataset from the same single original dataset, we create an environment where the potential benefit of transfer learning can be determined on real-world gene expression data. For this we utilized gene expression data provided as Reads Per Kilobase of transcript per Million mapped reads (RPKM) derived from over 1600 cells of the primary visual cortex of the adult mouse brain26. After the application of the pre-processing steps (see Supplementary Section 3.1. for details) the dataset contained expression levels of 9547 genes in 1658 cells.
We deemed this dataset to be of sufficient complexity in terms of taxonomic diversity (it contains 23 GABAergic neuronal, 19 glutamatergic neuronal and 7 non-neuronal cell types) and in terms of total cell count, to enable cluster-restricted subsampling and thus the application of transfer learning approaches.
We ran 100 repetitions splitting the data into a source dataset of 1000 cells and a target dataset of 650 cells each time, which is subsampled even further (to 25, 50, 100, 200, 400, 650 target cells) to assess performance for different sample sizes. To investigate the influence of complete and incomplete overlap between the clusters of source and target datasets, transcriptomic cell types assigned to either dataset were controlled. Complete overlap meant randomly assigning cells into source and target. Incomplete overlap is achieved by assigning the two largest clusters of the dataset (Glutamatergic L4 cells and GABAergic Pvalb cells) to be either an exclusive source or an exclusive target cluster, respectively. All other clusters are shared amongst both source and target in this setting.
The transfer learning approach and its baselines are now investigated under two different conditions. Firstly, we assume that no ground truth labels are available and generate labels for 18 cell clusters via NMF clustering73,74 on the whole dataset. We interpret this clustering, based as it is on the totality of the data, as a ground truth clustering and apply our method and the baseline algorithms to a subset of the dataset, to see how each method performs relative to this definition of ground truth when not all of the data is available. Secondly, we use the data driven clustering labels provided in the original paper and take those as the ground truth labels. Specifically, we use a cut-off in the provided clustering hierarchy that results in 18 clusters. Given those alternative ground truth labels, we once again run TargetCluster, ConcatenateCluster and TransferCluster. See Supplementary Section 3.3. for a more detailed description of the two different sets of ground truth labels.
Independent source and target dataset
As a real-world application of the transfer learning approach we analyze two entirely independent, but biologically related, datasets. To improve the clustering results of a relatively small target dataset from Hockley et al. 201821, we transfer knowledge from a larger source dataset from Usoskin et al. 201420 both derived from the rodent somatosensory system. The somatosensory system is responsible for detecting mechanical, thermal and chemical stimuli to which an organism can choose to elicit a behavioural response. Primary sensory neurons innervate the vast majority of internal hollow organs, joints, muscles and the skin evoking conscious sensation in the event of these stimuli. This is most clearly exemplified by pain in the case of potentially harmful or noxious stimuli, such as burning or cutting of the skin. In Usoskin et al, transcriptomic analysis of 622 primary sensory cell bodies, which reside within the dorsal root ganglia (DRG), reveals significant diversity in cell type (11 types) and sensitivity to a diverse range of stimuli modalities (e.g. thermosensitive, itch sensitive, nociceptive) to which an organism is exposed. However, previous retrograde tracing experiments show that only 5–10 % of DRG neurons project to internal (visceral) targets, such as the gastrointestinal tract, and as such are likely only represented by ~ 30–60 cells in the Usoskin et al. dataset. Such small cell numbers limit subtype assignment of cells in this organ. In order to overcome this limitation, scRNA-Seq has been performed on retrograde labeled DRG neurons known to selectively innervate the gastrointestinal tract (colonic DRG neurons), providing transcriptomic analysis of 314 cells from this specific organ that cluster into 7 distinct subtypes21. However, it is unclear whether de novo clustering of colonic DRG neurons identifies established clusters previously identified in larger datasets such as Usoskin et al. (hereafter designated ‘Usoskin’) or whether novel cell types exist within this dataset (hereafter designated ‘Hockley’).
After the application of cell and gene filter to the Hockley dataset provided as Transcripts Per Million (TPM), the number of genes decreased from 45513 to 9651; none of the 314 cells were filtered out. Pre-processing of the Usoskin dataset provided as Counts Per Million (CPM) left 9280 of the 20191 genes and 501 of the 622 cells. Please see Supplementary Section 4.1. for an investigation of the specific expression levels in these datasets and the parameter values that were consequently chosen for pre-processing. Since the methods require both source and target to have identical feature space, only the subset of genes that appear in both source and target data were used, leaving us with 4402 genes in both sets. In initial experiments, the original source and target data were used, however in later experiments, a batch effect removal approach was applied to control for the integration of single-cell transcriptomic data across different conditions and technologies. Here, we applied Seurat batch effect removal56 to combine the Hockley and the Usoskin data, and separated the result back into the original datasets which were then provided to TargetCluster, ConcatenateCluster and TransferCluster.
As an additional pre-processing step we investigated the effect of imputation on the clustering results. MAGIC78, a widely used method for imputing missing values to overcome zero-inflation in scRNA-Seq data, was applied to both datasets and the pre-processed datasets were then provided to the three methods under investigation.
Using either the original datasets or the pre-processed, batch effect removed or imputed datasets, the results of TargetCluster, ConcatenateCluster and TransferCluster were assessed in terms of performance via a comparison to the clustering of the original paper21, the evaluation of t-SNE plots and differentially expressed genes to determine putative cellular function to neuronal subtypes. Since SC3 is a non-convex method it yields different results for each run. In order to provide quantification of the robustness of the three methods, we applied each 1000 times and counted the number of times three key clusters of interest were successfully identified. These clusters were selected based on their biological relevance as described in the original paper, further details of which can be found in the results section.
Once again, experiments were run under two conditions. Firstly, we assumed that reliable source data labels are not available and we generated cell labels for the Usoskin dataset via NMF clustering. Secondly, we use labels from Usoskin et al. (generated via an iterative PCA approach). Usoskin et al. provide labels at three different levels of the hierarchy producing 4, 8 or 11 clusters. We investigate results based on all of those, calling them level 1, 2 and 3 labels, respectively. We also investigate a scenario where we generate the labels via NMF clustering instead of using the labels presented in Usoskin et al. Here, however, we only present results based on using level 3 labels from the original publication. Please see Supplementary Section 4.3 and 4.4 for a detailed description of the different sets of ground truth labels and the corresponding clustering results.
In order to assess whether rare cell types were present in the Hockley dataset, the number of clusters to group the cells of the target dataset in was chosen to be 7.




![Buildbox Free - How To Make 2D Platformer Game [PART 1]](https://www.danielparente.net/info/uploads/sites/3/2020/01/Buildbox-Free-How-To-Make-2D-Platformer-Game-PART-150x150.jpg)
